Functions/Subroutines | |
| subroutine, public | free_scattering (mo_integral_storage, molecular_orbital_basis, a, min_energy, max_energy, nE) |
| Performs the free potential scattering given the location of the transformed overlap and kinetic energy integrals, orbital basis set and R-matrix radius. More... | |
| subroutine | calc_rmat_e (rmatrix, e, a0, u, ek, n, n_chan) |
| subroutine | bessel (r, p, nu, nufrac, j, dj, y, dy) |
Function/Subroutine Documentation
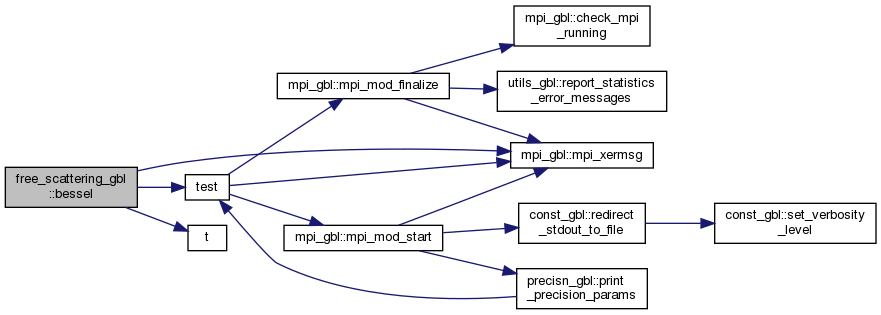
◆ bessel()
| subroutine free_scattering_gbl::bessel | ( | real(kind=cfp) | r, |
| real(kind=cfp) | p, | ||
| integer(kind=4) | nu, | ||
| real(kind=cfp) | nufrac, | ||
| real(kind=cfp) | j, | ||
| real(kind=cfp) | dj, | ||
| real(kind=cfp) | y, | ||
| real(kind=cfp) | dy | ||
| ) |
Here is the call graph for this function:
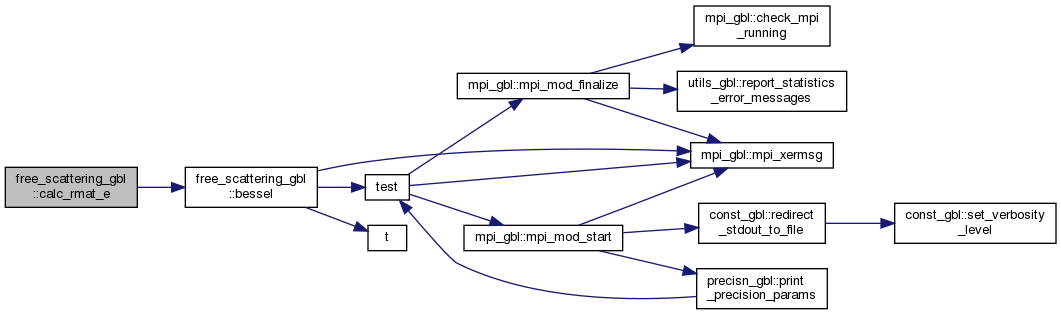
◆ calc_rmat_e()
| subroutine free_scattering_gbl::calc_rmat_e | ( | real(kind=cfp), dimension(:,:), intent(out) | rmatrix, |
| real(kind=cfp), intent(in) | e, | ||
| real(kind=cfp), intent(in) | a0, | ||
| real(kind=cfp), dimension(:,:), intent(in) | u, | ||
| real(kind=cfp), dimension(:), intent(in) | ek, | ||
| integer, intent(in) | n, | ||
| integer, intent(in) | n_chan | ||
| ) |
Here is the call graph for this function:
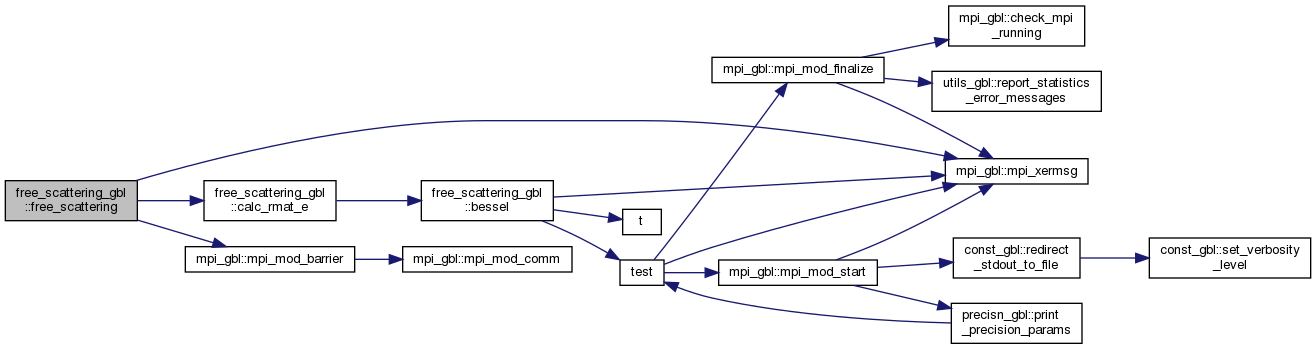
◆ free_scattering()
| subroutine, public free_scattering_gbl::free_scattering | ( | type(integral_storage_obj), intent(inout) | mo_integral_storage, |
| type(molecular_orbital_basis_obj), intent(inout) | molecular_orbital_basis, | ||
| real(kind=cfp), intent(in) | a, | ||
| real(kind=cfp), intent(in) | min_energy, | ||
| real(kind=cfp), intent(in) | max_energy, | ||
| integer, intent(in) | nE | ||
| ) |
Performs the free potential scattering given the location of the transformed overlap and kinetic energy integrals, orbital basis set and R-matrix radius.
Here is the call graph for this function: